eScience use case
1.1 Organisation behind the Case.
AGH University of Science and Technology in Kraków is one of the best Polish technical universities. It educates students in 54 branches of studies, including over 200 specializations run at 16 faculties and employing teaching and research staff of 1 887 persons (including 181 full professors). The Department of Computer Science, which participates in the project, employs teaching and research staff of over 80 people, devoting their research efforts to various IT directions, including scalable distributed systems, cross-domain computations in loosely coupled environments, knowledge management and support for life sciences.
1.2 Objectives
Within the scope of research projects, AGH collaborates closely with researchers and application users from the eScience domain, both local and international. The interesting use cases for PaaSage are those that require either large-scale workflow or data farming processing. AGH is either involved directly in supporting these applications on grids and clouds or develops tools that enable and facilitate execution of them on these infrastructures.
Local eScience applications and tools are related mostly to the PL-Grid project users and include:
- Bioinformatics applications, in collaboration with the Jagiellonian University Medical College.
- Investigating potential benefits of data farming application to study complex metallurgical processes, in collaborationFaculty of Metals Engineering and Industrial Computer Science AGH.
International collaborations in eScience domain include:
- Virtual Physiological Human initiative,
- Multiscale applications using workflow tools and MAPPER framework,
- Collaboration with Pegasus team from University of Southern California for support of scientific workflows on cloud infrastructures,
- Mission planning support in military applications with data farming within the EDA EUSAS project.
The two main tools that are developed by AGH to support these applications are:
- HyperFlow workflow execution engine that is based on hypermedia paradigm and supports flexible processing models such as data flow, control flow, and includes the support for large-scale scientific workflows which can be described as directed acyclic graphs of tasks.
- Scalarm is a massively self-scalable platform for data farming, which supports phases of data farming experiments, starting from parameter space generation, through simulation execution on heterogeneous computational infrastructure, to data collection and exploration.
1.3 Large-scale scientific workflows using HyperFLow
A current cloud-based workflow execution in HyperFlow (https://github.com/dice-cyfronet/hyperflow) is depicted in 1. When invoked, the function of a process sends a job specification to a remote message queue. It immediately subscribes to the queue in order to wait for job results. In parallel, local Execution Engines residing on Virtual Machines deployed in a cloud fetch the jobs from the queue, invoke the appropriate application components, and send the results back to the queue. When the results are received in the function of the Process, the callback is invoked. Note that multiple VMs and execution engines could be deployed and connected to the same queue which would act not only as a communication medium, but also as a load balancing mechanism.
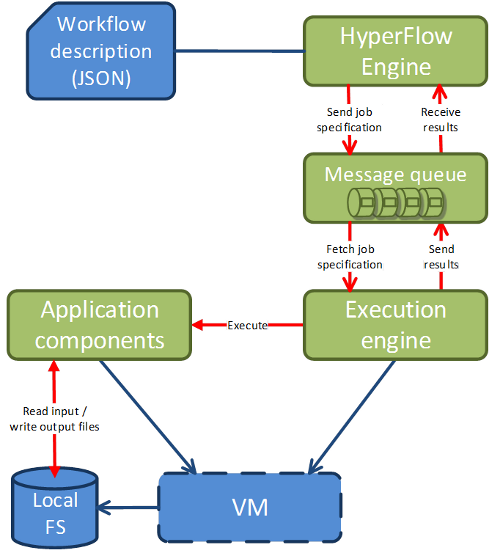
Figure: Workflow execution in a cloud enacted by the HyperFlow engine
Currently used mechanisms to deploy scientific workflows on clouds require deployment of a cluster of virtual machines for running the worker nodes that execute workflow tasks. This step can be done manually, or use some automated tools for infrastructure setup and application deployment.
We believe that integrating the HyperFlow engine with the PaaSage platform will have mutual benefits. HyperFlow will benefit from PaaSage cloud deployment, execution, and autoscaling capabilities. The PaaSage platform, in turn, will gain the capability to support a new class of applications: large-scale resource-intensive scientific workflows. This support will be in terms of composition of existing application components into workflows, and – in conjunction with the workflow scheduler – their effective autoscaling.
1.4 Conducting data farming experiments with Scalarm
Data farming is a methodology based on the idea that by repeatedly running a simulation model on a vast parameter space, enough output data can be gathered to provide an meaningful insight into relation of model’s properties and behaviours, with respect to simulation’s input parameters. Scalarm provides a complete platform for conducting data farming experiments with heterogeneous computational infrastructures. The user has at his disposal a set of well-known Design of Experiment methods for generating the experiment’s parameter space (as depicted in 2).
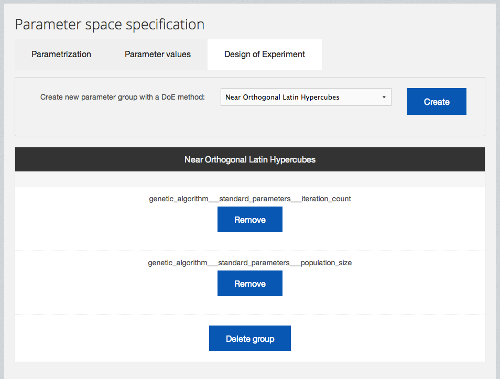
Figure : The parameter space definition dialog in Scalarm.
The progress of each data farming experiment can be monitored (as depicted in 3) and if required the initial parameter space can be extended. In addition, the user can dynamically increase the amount of computational resources dedicated to execute the experiment and the platform will scale itself accordingly to the new size of the experiment.
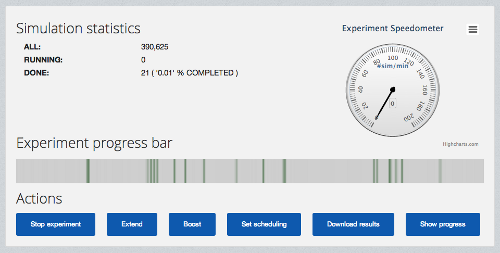
Figure : The progress monitoring view of a data farming experiment in Scalarm.
For more information about the Scalarm platform, please visit http://www.scalarm.com
1.4.1 Current status
The Scalarm architecture follows the master-worker design pattern, where the worker part is responsible for executing simulation, while the master part coordinates data farming experiments. Each part is implemented with a set of loosely coupled services. In the current version, the user manually manages resources of the worker part, i.e. additional workers are scheduled to different infrastructures manually. In regard to the master part, an administrator can define so called scaling rules to specify the scaling conditions and actions for each internal service.
1.4.2 Target picture
By integrating with the PaaSage infrastructure, we intend to provide a fully autonomous platform for data farming in regard to scalability. Deployment of a dedicated Scalarm instance on multicloud environments will be automated with PaaSage components. Furthermore, the scaling rules for each Scalarm service will be derived from user requirements automatically. On the other hand, PaaSage users will be capable to conduct data farming experiments without prior investments in either infrastructure or execution coordinating software required.

